UML-08: Anomaly Detection
Learning Objectives
After reading this post, you will be able to:
- Understand the three main approaches to anomaly detection
- Implement Isolation Forest and Local Outlier Factor
- Know when to use each anomaly detection method
- Handle contamination rate and threshold selection
Theory
The Intuition: The Rare Species Hunter
Imagine you are a biologist exploring a new island.
- Normal Data (Sparrows): You see thousands of small, brown birds. They are everywhere and look mostly the same.
- Anomalies (The Phoenix): Suddenly, you see a giant, glowing red bird. It stands out immediately.
Anomaly Detection is the set of algorithms designed to find these “Rare Species” in a sea of common data, without knowing beforehand what they look like.
Applications:
- 🏦 Fraud detection (unusual transactions)
- 🖥️ System monitoring (server failures)
- 🏭 Quality control (defective products)
- 🏥 Medical diagnosis (rare diseases)
Three Approaches to Anomaly Detection
| Approach | Method | Assumption |
|---|---|---|
| Statistical | Z-score, IQR | Data follows known distribution |
| Distance-based | LOF, k-NN | Anomalies are far from neighbors |
| Tree-based | Isolation Forest | Anomalies are easier to isolate |
Statistical Methods
Z-Score
Points with $|z| > 3$ are often considered anomalies:
$$z = \frac{x - \mu}{\sigma}$$
Interquartile Range (IQR)
Points outside the “fences” are outliers:
- Lower fence: $Q_1 - 1.5 \times IQR$
- Upper fence: $Q_3 + 1.5 \times IQR$
The “Square Peg” Problem (Why Z-Score Fails)
Z-score looks at features individually.
- Imagine a person who is 6'0" (Normal).
- Imagine a person who weighs 100 lbs (Normal).
- But a person who is 6'0" AND 100 lbs? That’s an anomaly! Z-score might miss this because each dimension alone looks fine. It can’t see the relationship between features.
Isolation Forest: The “20 Questions” Game
Think of this as playing “20 Questions” to identify an animal.
- To identify a Sparrow (Standard Point): You need many questions. “Is it small? Yes. Brown? Yes. Beak shape? Conical…” because so many birds fit this description.
- To identify a Phoenix (Anomaly): You need only one question. “Is it on fire? Yes.” -> Found it!
Isolation Forest builds random decision trees.
- Anomalies are distinct, so they get “isolated” near the root of the tree (few splits).
- Normal points are clustered together, so they end up deep in the leaves (many splits).
Why it works: The “Outsider” Principle
Because anomalies are " different" and “few”, random cuts are very likely to separate them from the rest of the data early on.
- Normal points are crowded together. You have to peel away many layers (cuts) to isolate one specific normal point.
- Anomalies sit alone. One or two random cuts are usually enough to fence them off.
Algorithm:
- Build many random trees (random splits on random features)
- Anomalies require fewer splits to be isolated
- Score = average path length across all trees
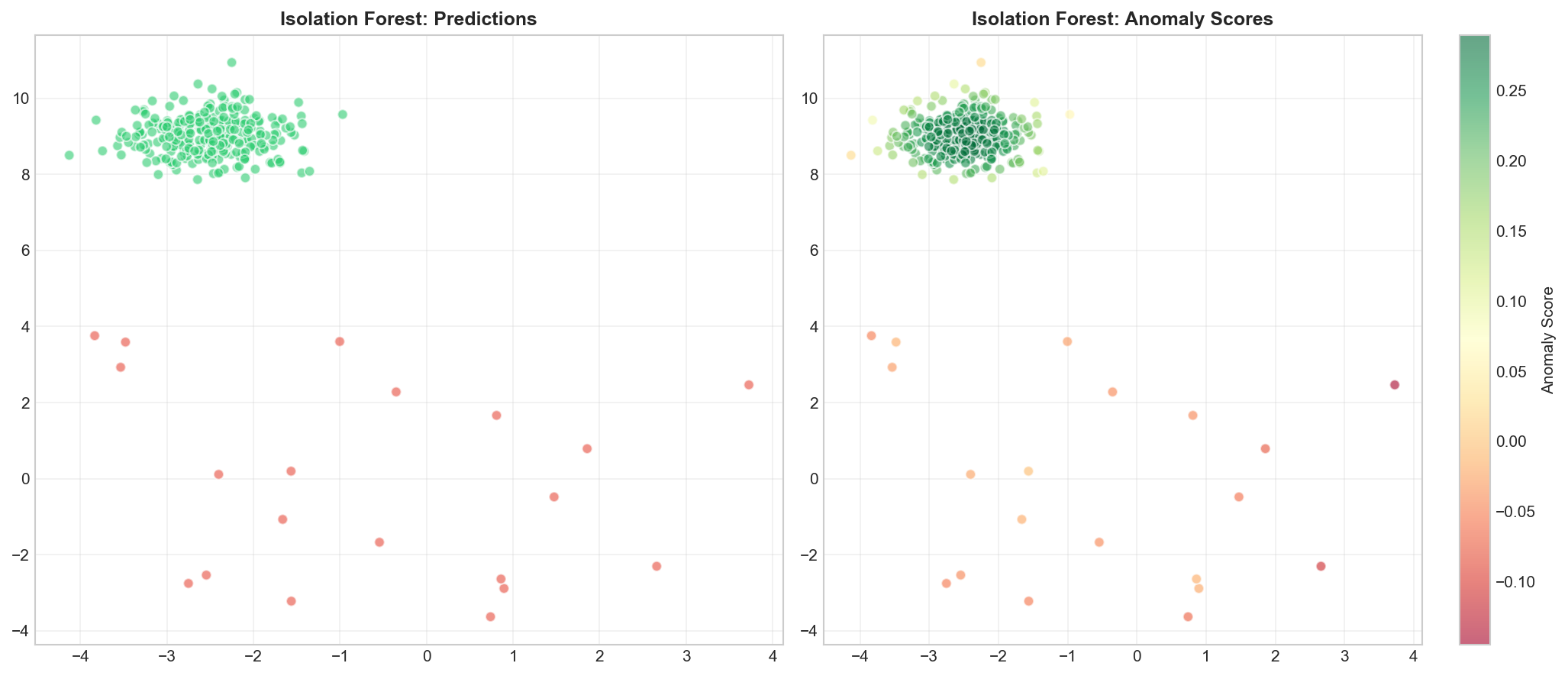
Anomaly Score: $$s(x, n) = 2^{-\frac{E[h(x)]}{c(n)}}$$
where $h(x)$ = path length, $c(n)$ = average path length in random trees.
- Score ≈ 1: anomaly
- Score ≈ 0: normal
- Score ≈ 0.5: uncertain
Local Outlier Factor (LOF): The “Loneliness” Index
LOF measures how isolated a point is compared to its neighbors.
- The Crowd (High Density): A penguin in a flock is happy because its neighbors are just as close to each other as they are to him.
- The Loner (Low Density): A penguin alone on an iceberg is lonely. His density is low, while his nearest neighbors (far away in the flock) have high density.
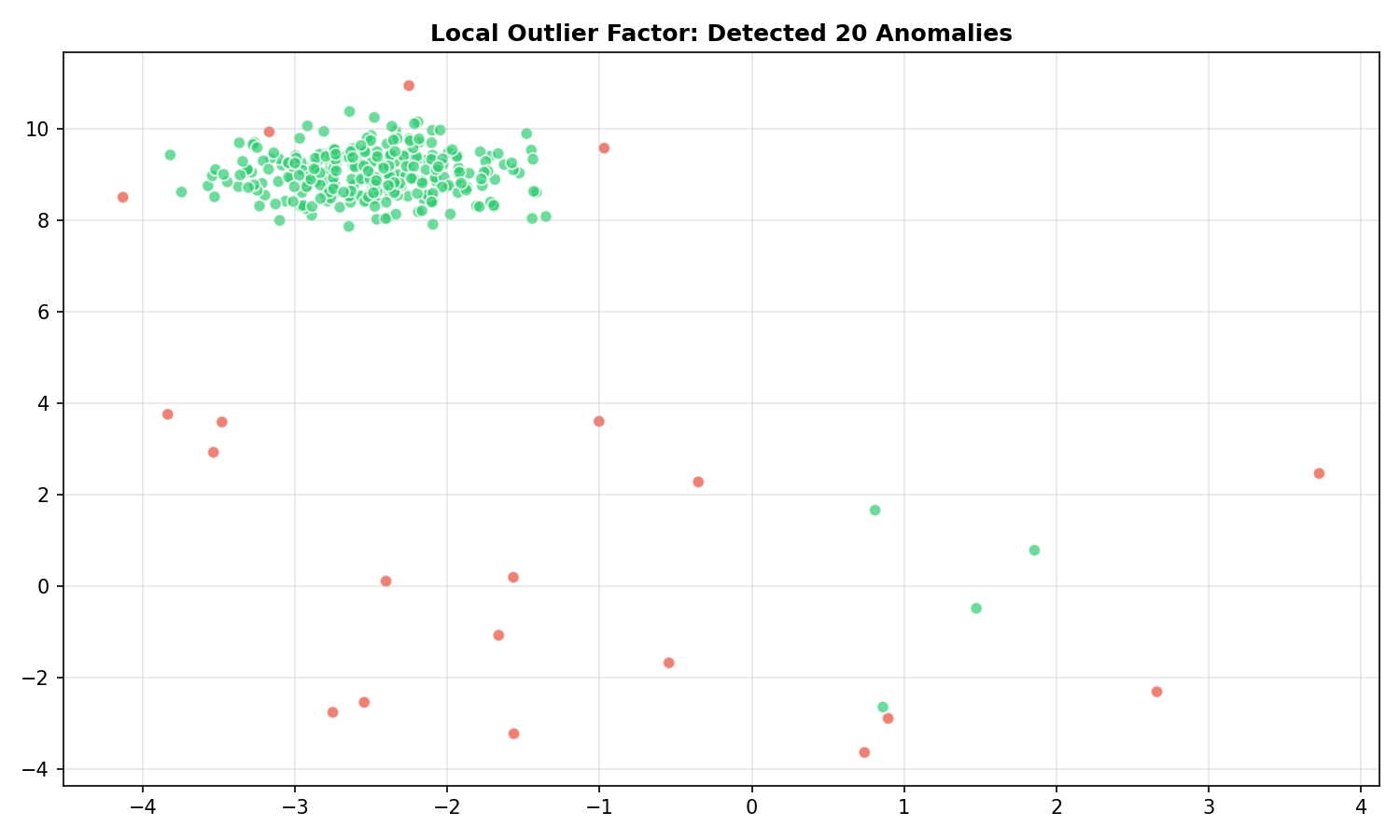
LOF » 1 means the point is in a lower-density region than its neighbors (Anomaly).
$$LOF(x) = \frac{\text{avg neighbor density}}{\text{density at } x}$$
- LOF ≈ 1: similar density to neighbors (normal)
- LOF » 1: lower density than neighbors (anomaly)
Why LOF? The “Density Problem” Imagine two clusters:
- City (High Density): Points are packed tight. An anomaly here might be just a block away.
- Countryside (Low Density): Points represent farms miles apart.
A global method (like k-NN distance) might flag all the farms as anomalies because they are far apart. LOF adapts: It knows that being 1 mile apart is “normal” in the countryside, but “anomalous” in the city.
Code Practice
Statistical Anomaly Detection
🐍 Python
| |
Output:
Isolation Forest
🐍 Python
| |
Output:
Visualizing Isolation Forest
🐍 Python
| |

Local Outlier Factor
🐍 Python
| |
Output:

Comparing Methods
🐍 Python
| |
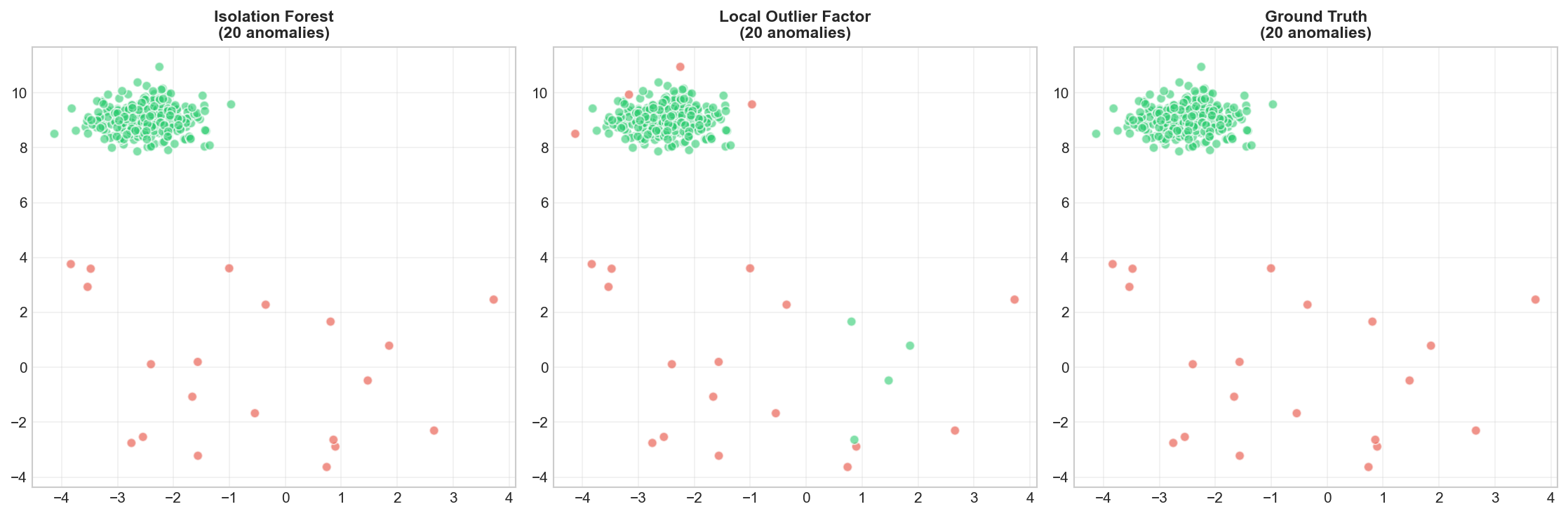
Deep Dive
Choosing the Right Method
| Method | Best For | Limitations |
|---|---|---|
| Z-score/IQR | Univariate, Gaussian data | Assumes distribution |
| Isolation Forest | High-dimensional, fast | Global outliers |
| LOF | Local outliers, varied densities | Slower, needs k choice |
| One-Class SVM | Complex boundaries | Scaling, parameters |
Handling Contamination Rate
The contamination parameter is crucial but often unknown:
- If known: set explicitly (e.g., 5% fraud rate)
- If unknown: try multiple values, use domain knowledge
- Alternative: use
autoand threshold scores manually
Frequently Asked Questions
Q1: How do I evaluate anomaly detection without labels?
Difficult! Options:
- Manual inspection of flagged anomalies
- Domain expert validation
- If partial labels exist: precision/recall on known anomalies
Q2: What if I have labeled anomalies?
Then you have a supervised classification problem! Consider:
- One-Class SVM (trained on normals only)
- Supervised classifiers with class imbalance handling
Q3: How do I handle high-dimensional data?
- Use Isolation Forest (handles high-D well)
- Apply PCA first, then anomaly detection
- Use distance metrics designed for high-D (e.g., cosine)
Summary
| Concept | Key Points |
|---|---|
| Anomaly | Unusual observation, differs from majority |
| Isolation Forest | Tree-based, isolates anomalies quickly |
| LOF | Density-based, compares to local neighborhood |
| Contamination | Expected proportion of anomalies |
| Evaluation | Difficult without labels; use domain knowledge |
References
- Liu, F.T. et al. (2008). “Isolation Forest”
- Breunig, M.M. et al. (2000). “LOF: Identifying Density-Based Local Outliers”
- sklearn Outlier Detection