UML-03: Hierarchical Clustering
Learning Objectives
After reading this post, you will be able to:
- Understand the difference between agglomerative and divisive clustering
- Master the four main linkage methods and when to use each
- Read and interpret dendrograms for data exploration
- Choose the optimal number of clusters using dendrogram analysis
Theory
The Intuition: The Digital Librarian
Imagine you have a messy hard drive with thousands of random files (images, documents, code). You don’t just dump them into 3 arbitrary buckets. Instead, you organize them hierarchically:
- Files go into specific sub-folders (e.g., “vacation_photos”).
- Sub-folders go into broader categories (e.g., “Photos”).
- Categories go into the root drive (e.g., “D: Drive”).
This is exactly what Hierarchical Clustering does. It organizes your data into a tree of nested clusters, giving you a complete map of how data points relate to each other at different levels of granularity.
What is Hierarchical Clustering?
Unlike K-Means, which forces you to choose $K$ clusters upfront (flat clustering), Hierarchical Clustering builds a tree structure (Dendrogram) that preserves the relationships between all points.
Two main approaches exist:
| Approach | Direction | Strategy |
|---|---|---|
| Agglomerative (bottom-up) | ⬆️ Merge | Start with N clusters, merge similar ones |
| Divisive (top-down) | ⬇️ Split | Start with 1 cluster, split into smaller ones |
Agglomerative Clustering Algorithm
The algorithm proceeds as follows:
- Initialize: Each data point is its own cluster ($N$ clusters)
- Find closest pair: Identify the two most similar clusters
- Merge: Combine them into a single cluster
- Repeat: Until only one cluster remains
- Output: A hierarchical tree (dendrogram)
Linkage: The Rules of Organization
The librarian needs a rule to decide which folders to merge next. How do we measure distance between clusters?
Different linkage methods define cluster distance differently:
| Linkage | Nickname | Distance Definition | Characteristics |
|---|---|---|---|
| Single | “The Optimist” | Min distance (closest points) | Creates long, chain-like clusters. Good for non-globular shapes. |
| Complete | “The Skeptic” | Max distance (farthest points) | Creates compact, spherical bubbles. Sensitive to outliers. |
| Average | “The Democrat” | Average of all pairs | Balanced, but affected by noise. |
| Ward | “The Architect” | Minimize variance increase | Creates similarly sized, spherical clusters. Default & usually best. |
Mathematical Definitions
For clusters $A$ and $B$:
Single linkage: $$d(A, B) = \min_{a \in A, b \in B} |a - b|$$
Complete linkage: $$d(A, B) = \max_{a \in A, b \in B} |a - b|$$
Average linkage: $$d(A, B) = \frac{1}{|A||B|} \sum_{a \in A} \sum_{b \in B} |a - b|$$
Ward’s method: $$d(A, B) = \sqrt{\frac{2|A||B|}{|A|+|B|}} |\mu_A - \mu_B|$$
The Dendrogram: The Map of Your Data
A dendrogram is a tree diagram showing the full history of merges or splits. It is essentially the “directory tree” of your data.
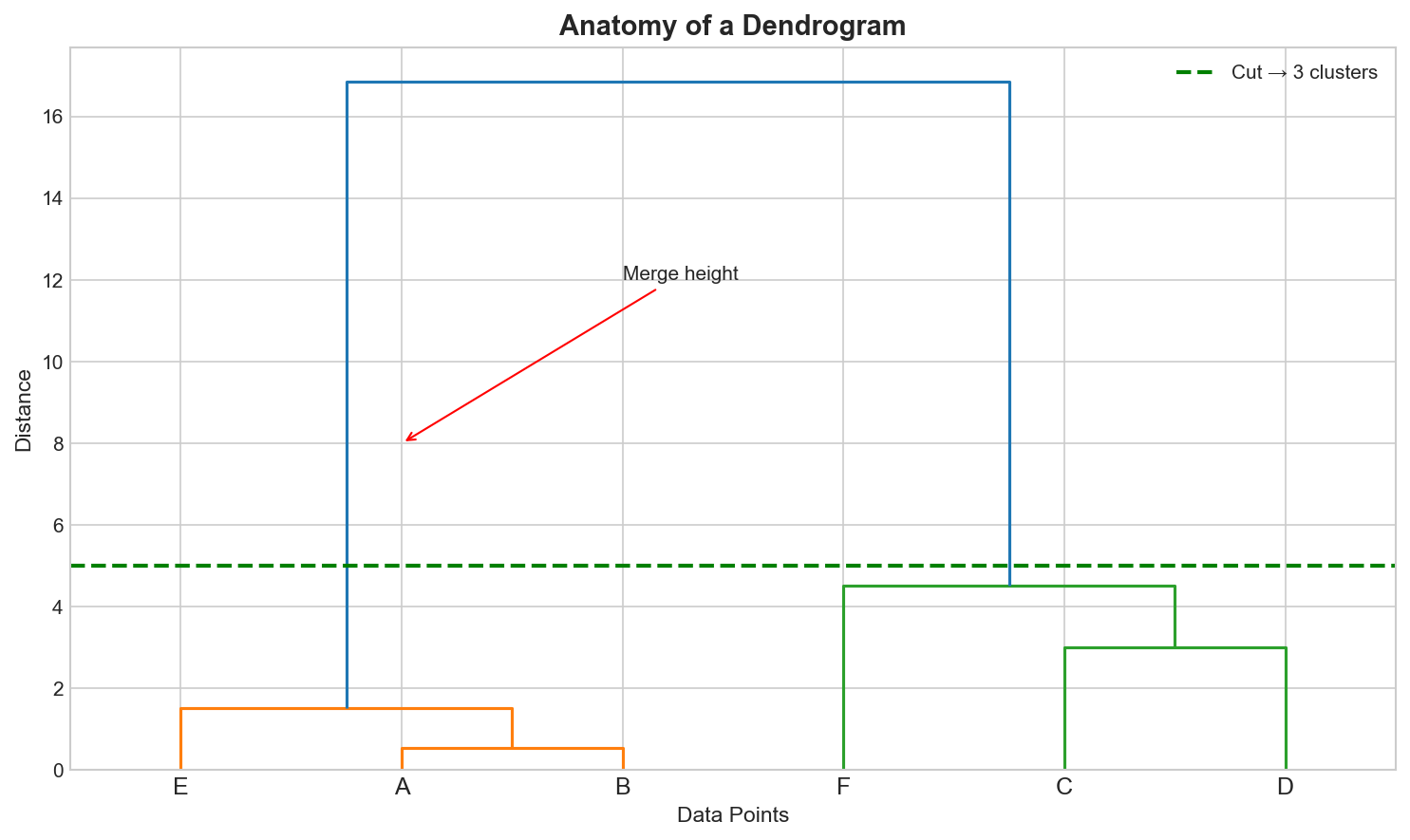
Key elements:
- Leaves: Individual data points (bottom)
- Vertical lines: Clusters being merged
- Height: Distance at which merge occurs
- Horizontal cut: Determines number of clusters
To get $K$ clusters, draw a horizontal line that crosses exactly $K$ vertical lines.
Choosing the Number of Clusters
Unlike K-Means, hierarchical clustering doesn’t require specifying $K$ upfront. Two approaches:
1. Visual inspection of dendrogram
- Look for large gaps in merge heights
- Cut where there’s the biggest “jump”
2. Inconsistency coefficient
- Compare each merge height to previous merges
- High inconsistency suggests a natural boundary
Code Practice
Now, let’s hand over the job to the algorithm. We will let Python act as the digital librarian, automatically sorting our raw data into a structured hierarchy.
Agglomerative Clustering with sklearn
🐍 Python
| |
Output:
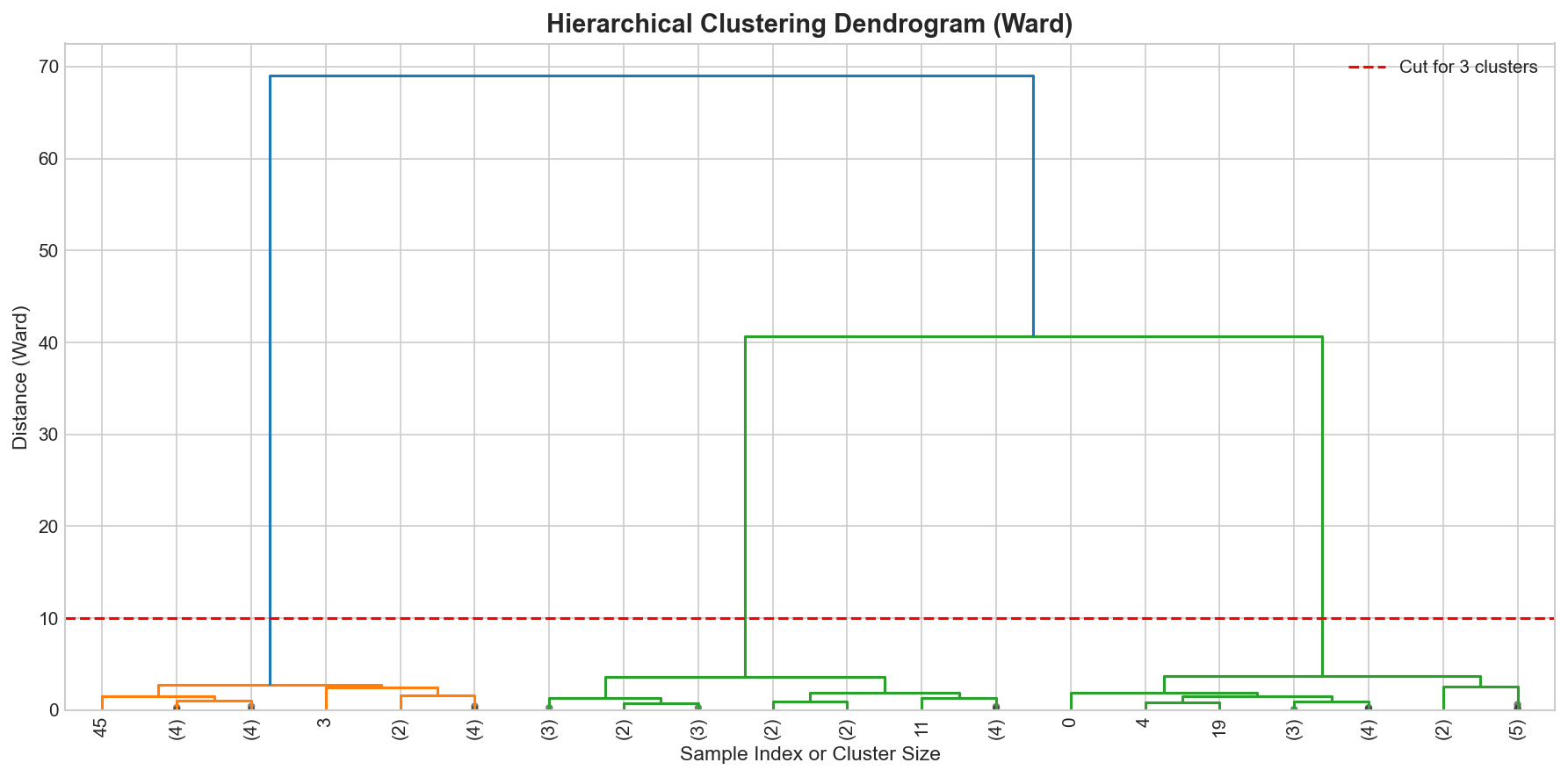
Creating and Interpreting Dendrograms
🐍 Python
| |
Output:
| |
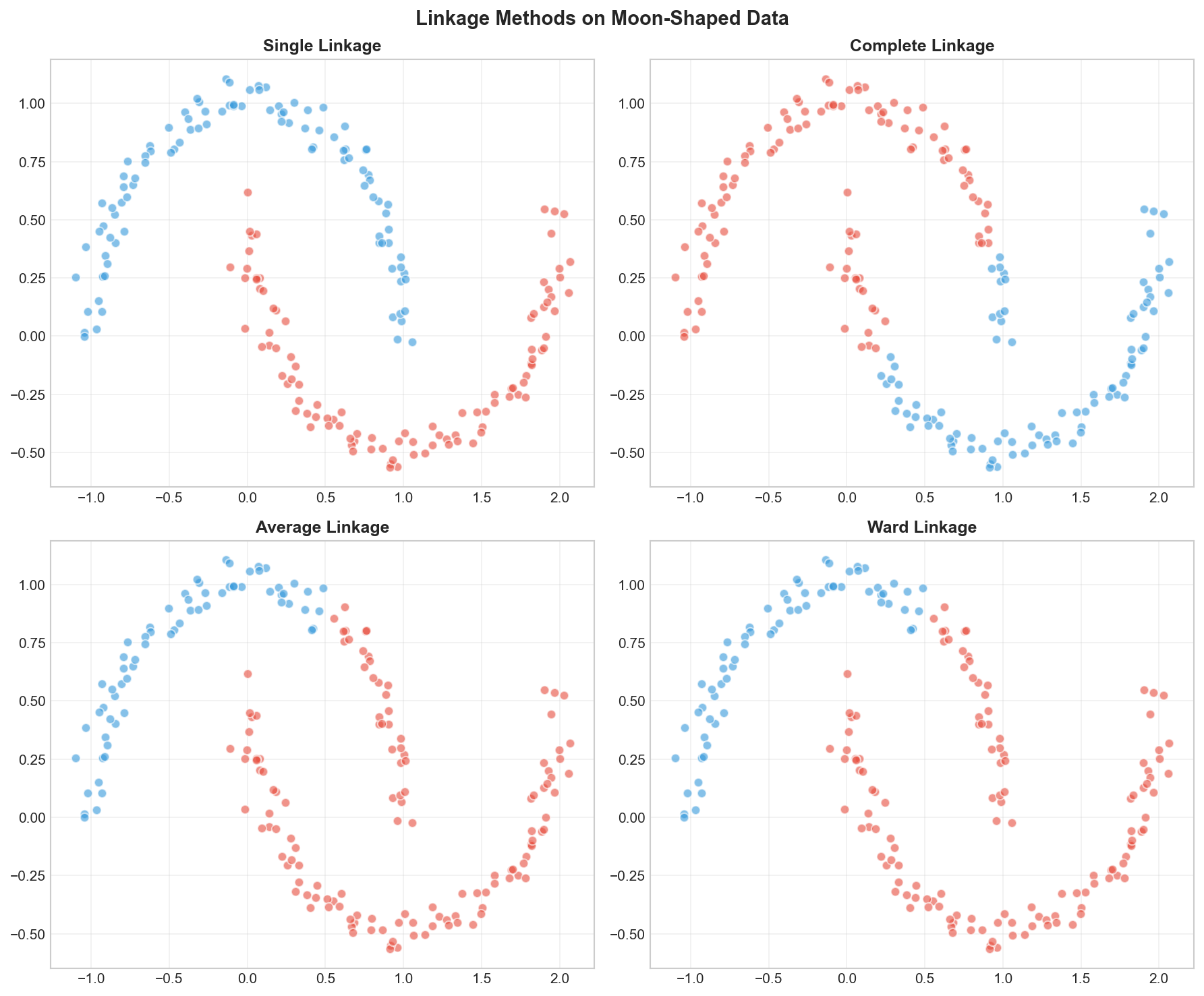
Comparing Linkage Methods
🐍 Python
| |

When to Use Which Linkage?
🐍 Python
| |
Output:
Interpreting the Results
Notice the stark difference!
- Single Linkage (“The Optimist”) achieved a perfect score (ARI=1.0) because it could “chain” along the curved moon shapes. It only needs one pair of close points to merge clusters.
- Ward (“The Architect”) failed (ARI ~0.45) because it tries to minimize variance, forcing the data into spherical (ball-like) shapes. It cut right through the crescents!
Decision Guide
So, which one should you choose?
| Method | Strength | Best For… |
|---|---|---|
| Ward | Balanced, cohesive clusters | General purpose. This is your default choice. Works best for noisy data or when you want clusters of similar sizes. |
| Single | Follows continuous shapes | Non-globular data (like the moons above) or detecting outliers. Warning: suffers from the “chaining” effect on noisy data. |
| Complete | Compact clusters | When you want to ensure the diameter of clusters is small (all points are somewhat similar). |
| Average | Compromise | A middle ground. Use when Ward is too strict about spherical shapes and Single is too loose. |
Deep Dive
Hierarchical vs K-Means
| Aspect | Hierarchical | K-Means |
|---|---|---|
| Specify K upfront | No | Yes |
| Dendrogram | Yes | No |
| Complexity | $O(N^2 \log N)$ to $O(N^3)$ | $O(N \cdot K \cdot d \cdot I)$ |
| Cluster shapes | Flexible (with single linkage) | Spherical |
| Scalability | Poor for large N | Good |
| Reproducibility | Deterministic | Random (depends on init) |
Computational Complexity
| Operation | Complexity |
|---|---|
| Distance matrix | $O(N^2)$ |
| Single/Complete linkage | $O(N^2 \log N)$ |
| Average/Ward linkage | $O(N^2 \log N)$ to $O(N^3)$ |
| Memory | $O(N^2)$ |
Scalability warning: Storing the $N \times N$ distance matrix becomes impractical for large datasets (e.g., 100K+ points). For big data, consider:
- Mini-batch approaches
- BIRCH (Balanced Iterative Reducing and Clustering using Hierarchies)
- K-Means first, then hierarchical on cluster centers
Frequently Asked Questions
Q1: When should I use hierarchical clustering over K-Means?
Use hierarchical clustering when:
- You don’t know how many clusters exist
- You want to explore data structure at multiple scales
- You need a dendrogram for interpretation
- Dataset is small enough ($N < 10000$)
Q2: Can I use hierarchical clustering with categorical data?
Yes, but you need an appropriate distance metric:
- Hamming distance: Count of differing attributes
- Jaccard distance: For binary/set data
- Gower distance: For mixed data types
Summary
| Concept | Key Points |
|---|---|
| Hierarchical Clustering | Builds tree of clusters without specifying K |
| Agglomerative | Bottom-up: merge similar clusters |
| Linkage Methods | Single, Complete, Average, Ward |
| Dendrogram | Visual representation of cluster hierarchy |
| Choosing K | Cut dendrogram where big gaps appear |
| Trade-off | More interpretable, but $O(N^2)$ complexity |
References
- sklearn Hierarchical Clustering
- scipy.cluster.hierarchy Documentation
- Müllner, D. (2011). “Modern hierarchical, agglomerative clustering algorithms”
- “The Elements of Statistical Learning” - Chapter 14.3