UML-04: DBSCAN and Density-Based Clustering
Learning Objectives
After reading this post, you will be able to:
- Understand the DBSCAN algorithm and its density-based approach
- Distinguish between core points, border points, and noise
- Tune the key parameters: epsilon (ε) and min_samples
- Know when DBSCAN outperforms K-Means and hierarchical clustering
Theory
The Intuition: The Archipelago
Imagine you are an explorer mapping an unknown ocean. You aren’t looking for perfectly round planets (like K-Means does). You are looking for archipelagos (chains of islands).
- Some islands are crescent-shaped.
- Some are long and thin.
- The open ocean (noise) should be ignored.
DBSCAN offers a way to navigate this: if you can step from one piece of land to another without swimming too far, they belong to the same island.
Why Density-Based?
Traditional algorithms like K-Means assume clusters are spherical “blobs”. But real-world data is often messy.
Core Idea: Density Connectivity
DBSCAN defines clusters based on local density:
- Dense regions = clusters
- Sparse regions = noise or boundaries
Two key parameters:
- ε (epsilon): Neighborhood radius
- min_samples: Minimum points to form a dense region
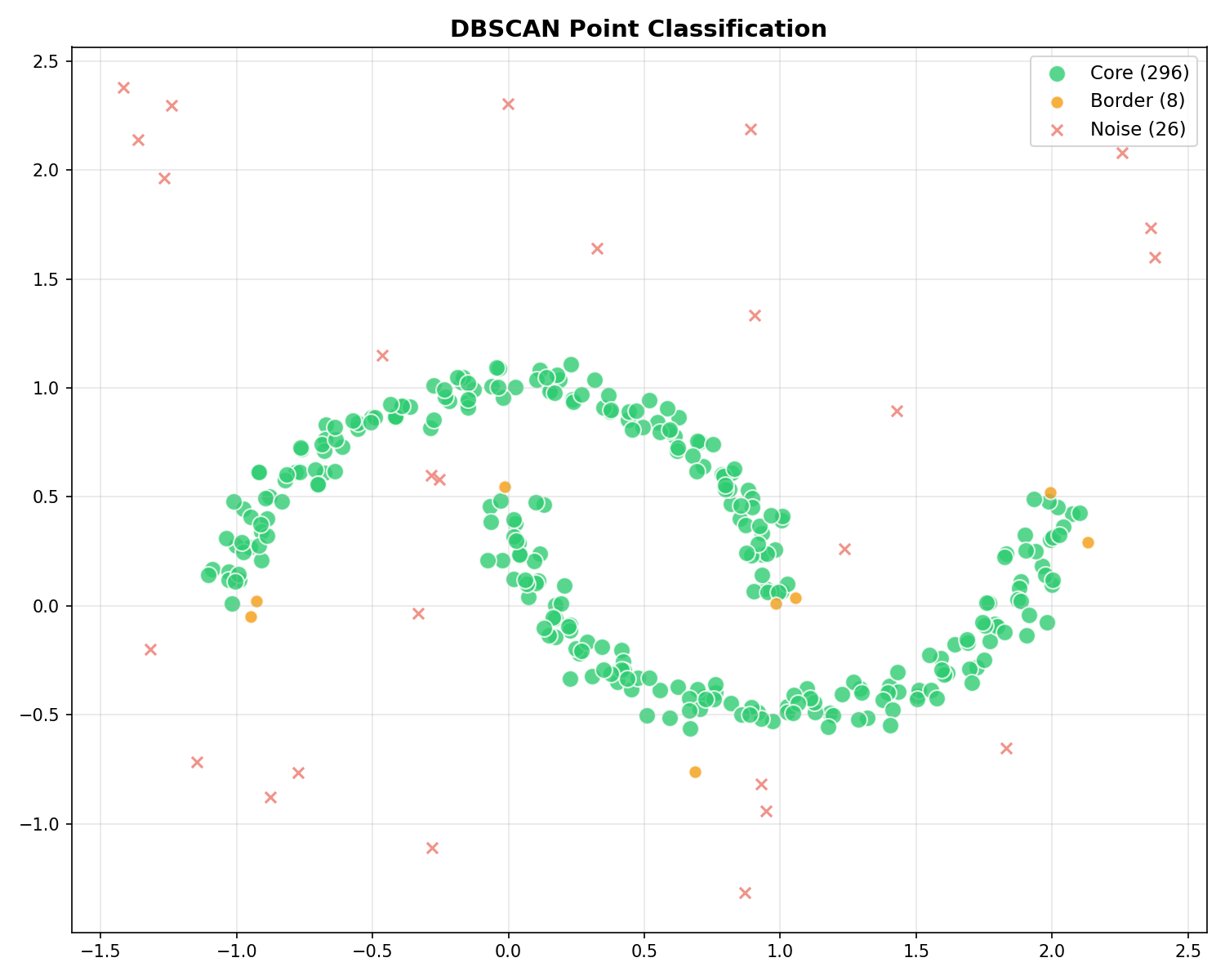
Point Classification
DBSCAN classifies every point into one of three types:
| Type | Analogy | Definition | Role |
|---|---|---|---|
| Core | Inland | Has ≥ min_samples within ε | Dense interior of the cluster. |
| Border | Beach | Reachable from Core, but < min_samples | Edge of the cluster. Part of the island, but thinner. |
| Noise | Ocean | Not reachable from any Core | Outliers / Water. Ignored. |
The DBSCAN Algorithm
Step-by-Step Narrative: How the Fire Spreads
- Ignition: We pick a random point. If it has enough neighbors (dense inland), we light a “signal fire” (Start a new cluster).
- Expansion: The fire spreads to all neighbors within reaching distance ($\epsilon$).
- Chain Reaction: If a neighbor is also a core point, it passes the fire to its neighbors. The cluster grows until the fire hits the water (border points) or runs out of fuel.
- Next Island: Once the fire dies out, we move to the next unvisited point and try to start a new fire.
- Water: If a point has no neighbors, it’s marked as water (noise) and ignored… for now. Usefull logic might claim it later!
Choosing Parameters
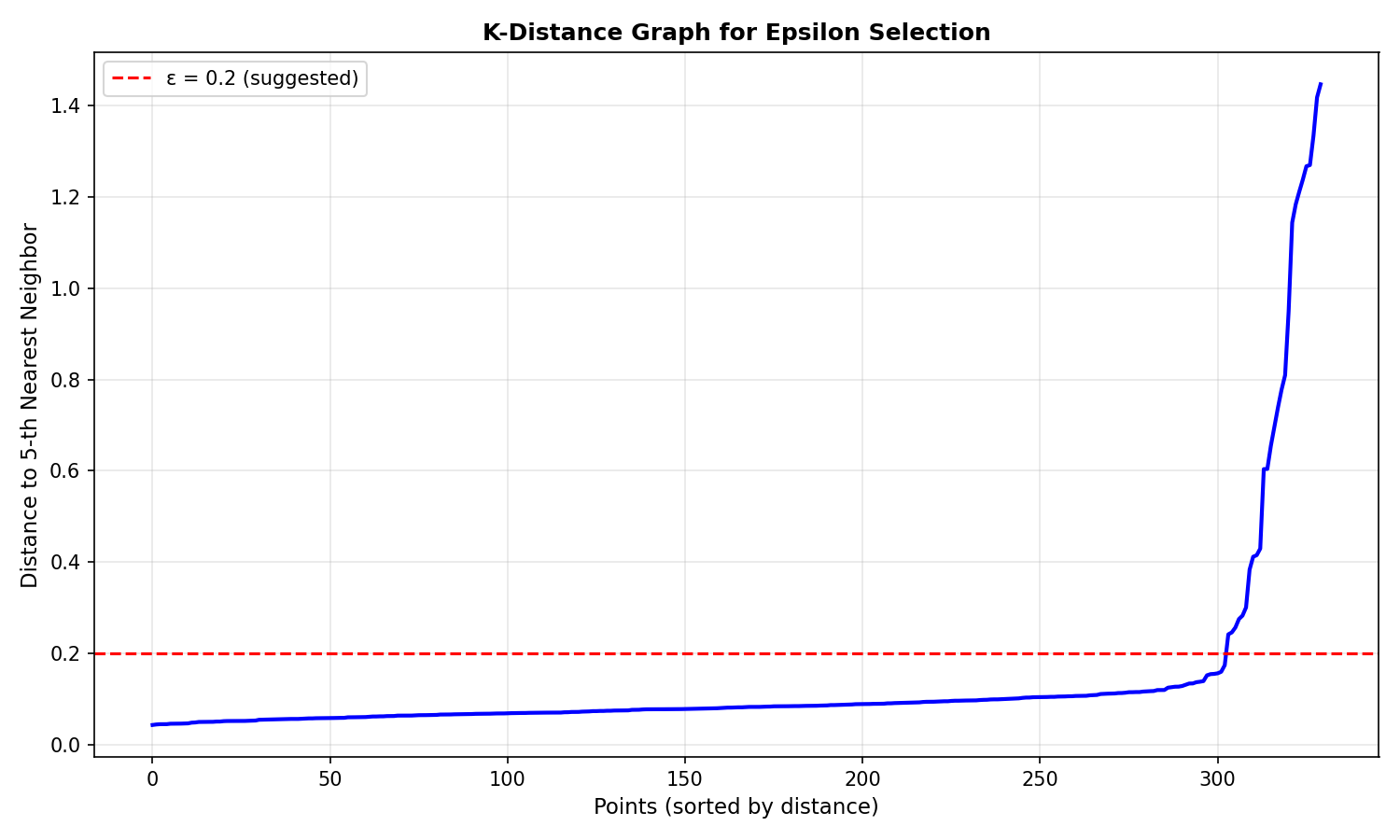
Epsilon ($\epsilon$): The Reach
Think of $\epsilon$ as your “arm span” or “swimming distance”.
- Too Small: You can’t reach the next person. Everyone becomes an isolated island (Noise).
- Too Large: You reach everyone, even those far away. The whole ocean becomes one giant country.
How to choose $\epsilon$? Use the K-distance graph: Calculate the distance to the $k$-th nearest neighbor for every point, sort them, and look for the “elbow” (where the curve sharply rises). That’s your optimal $\epsilon$.
min_samples: The Island Size
This defines how much land you need to officially call it an “island”.
- Small (e.g., 3): Even a few rocks count as an island. You’ll find many small, jagged clusters. Risk: False positives (noise usually looks like small islands).
- Large (e.g., 20): You only care about massive continents. Small details are washed away into the ocean. Risk: Losing small but valid clusters.
| min_samples | Effect |
|---|---|
| Low (3-4) | Many clusters. Good for cleaning distinct but small groups. |
| High (10+) | Few clusters. “Smoother” results, but small features vanish. |
Code Practice
DBSCAN on Moon-Shaped Data
🐍 Python
| |
Output:
DBSCAN vs K-Means Comparison
🐍 Python
| |
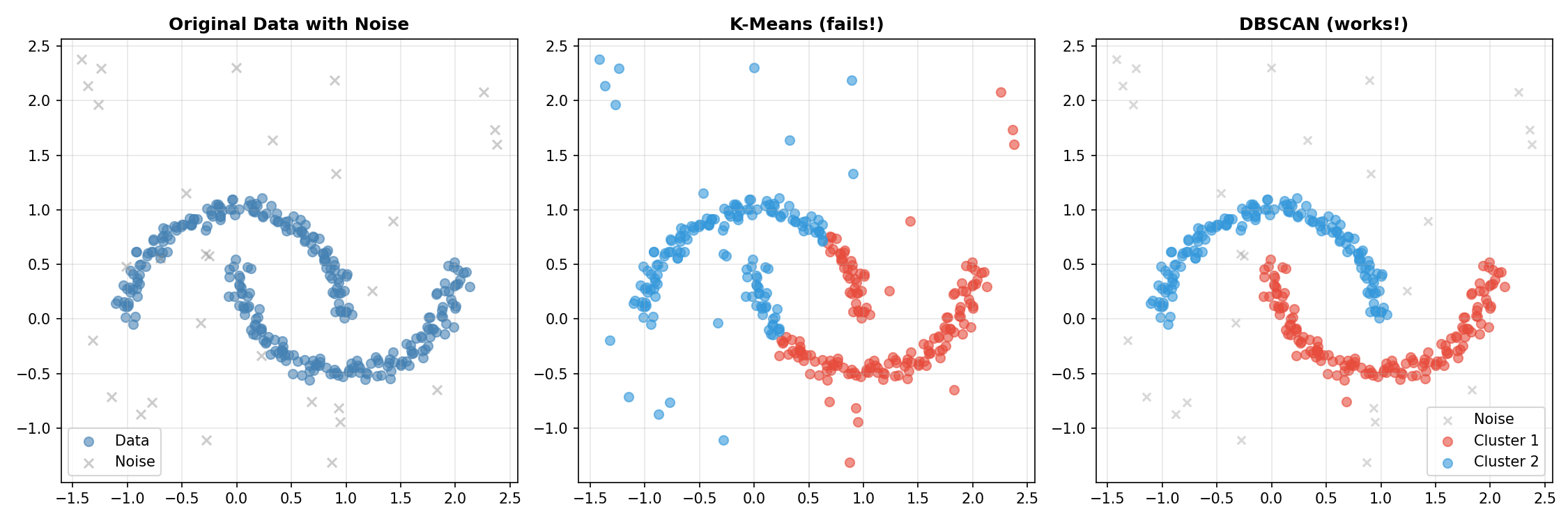
Interpreting the Results
Why did K-Means fail so badly here?
- K-Means searches for a center (centroid) and assumes the cluster spreads out spherically from there. It’s like trying to cover a banana with a circle—you’ll inevitably include empty space or cut the banana in half.
- DBSCAN doesn’t care about centers. It just checks: “Is there a neighbor nearby?” It crawled along the curve of the moon, point by point, perfectly tracing the shape. It also correctly ignored the random “noise” points because they didn’t have enough neighbors to start a cluster.
Finding Optimal Epsilon
🐍 Python
| |
Visualizing Core, Border, and Noise Points
🐍 Python
| |

Deep Dive
DBSCAN vs Other Methods
| Aspect | DBSCAN | K-Means | Hierarchical |
|---|---|---|---|
| Specify K | No | Yes | No |
| Cluster shapes | Any | Spherical | Depends on linkage |
| Noise handling | Built-in | None | None |
| Varying density | Struggles | OK | OK |
| Complexity | $O(N^2)$ or $O(N \log N)$ | $O(NKdI)$ | $O(N^2 \log N)$ |
When DBSCAN Struggles
DBSCAN limitations:
- Varying densities: Single ε can’t handle clusters with different densities
- High dimensions: Distance becomes less meaningful (“curse of dimensionality”)
- Parameter sensitivity: Results heavily depend on ε and min_samples choices
HDBSCAN: The Improved Version
HDBSCAN (Hierarchical DBSCAN) addresses varying density issues:
- Automatically finds clusters of different densities
- Only requires min_samples parameter
- More robust to parameter choices
Frequently Asked Questions
Q1: How do I choose epsilon if there’s no clear elbow?
Try multiple values and evaluate using:
- Silhouette score (for clusters without noise)
- Visual inspection of results
- Domain knowledge about expected cluster sizes
Q2: Why does DBSCAN give label -1?
Label -1 means noise — points that don’t belong to any cluster. This is a feature, not a bug! To reduce noise points, increase ε or decrease min_samples.
Q3: Can DBSCAN work with categorical data?
Not directly. Options:
- Use appropriate distance metrics (e.g., Gower distance)
- Convert to numerical encoding
- Use custom distance functions with sklearn’s DBSCAN
Summary
| Concept | Key Points |
|---|---|
| DBSCAN | Density-based clustering that finds arbitrary shapes |
| Parameters | ε (neighborhood radius), min_samples |
| Point types | Core (dense), Border (near core), Noise (-1) |
| Epsilon selection | Use k-distance graph, look for elbow |
| Strength | Handles noise, no K needed, any shape |
| Weakness | Struggles with varying densities |
References
- sklearn DBSCAN Documentation
- Ester, M. et al. (1996). “A Density-Based Algorithm for Discovering Clusters”
- Campello, R.J.G.B. et al. (2013). “Density-Based Clustering Based on Hierarchical Density Estimates” (HDBSCAN)
- HDBSCAN Documentation