ML-12: SVM: Kernel Methods and Soft Margin
Learning Objectives
- Understand the kernel trick
- Use polynomial and RBF kernels
- Implement soft-margin SVM
- Tune hyperparameter C
Theory
The Problem: When Linear Isn’t Enough
In last blog, hard-margin SVM was introduced to find linear decision boundaries. But what if data looks like this?
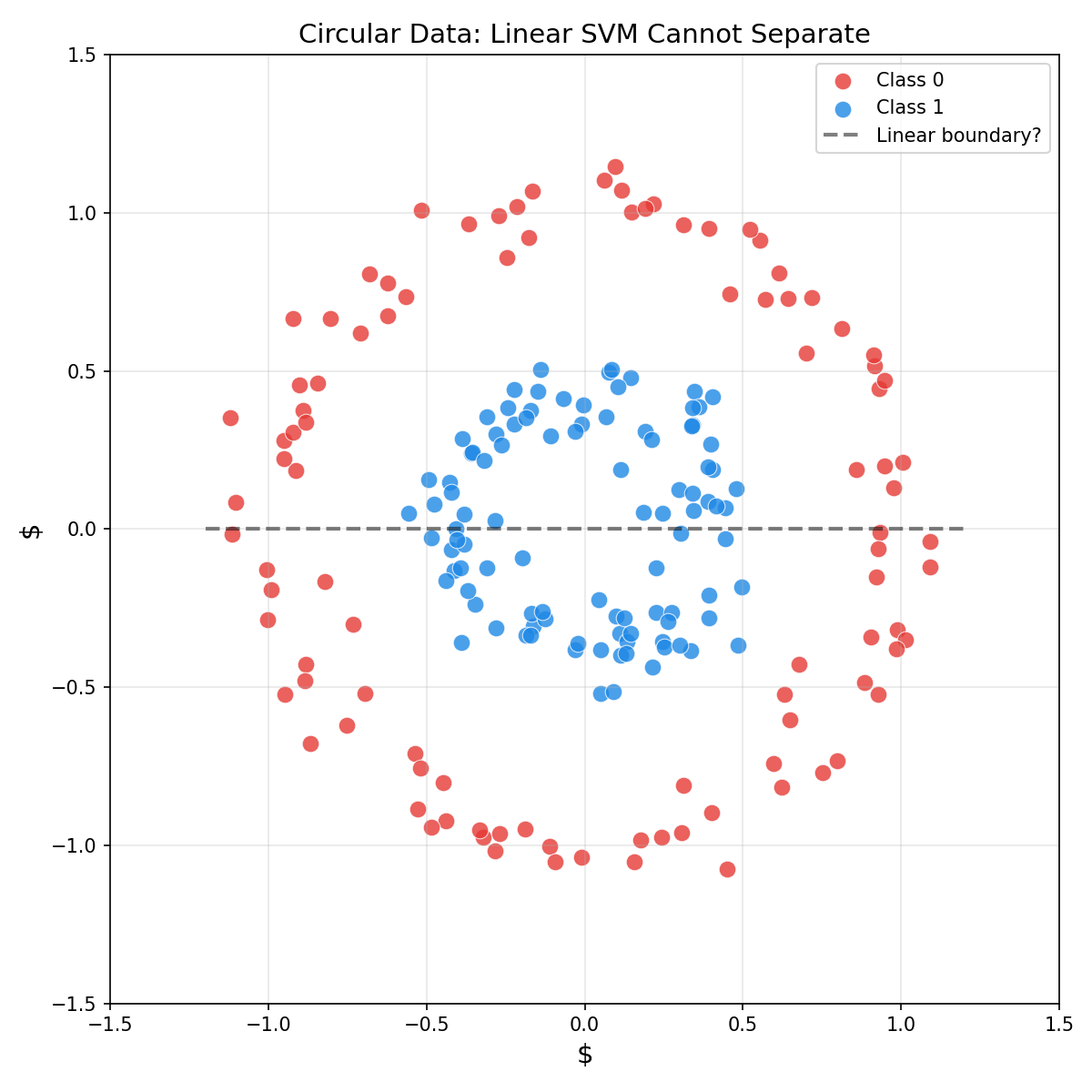
For data with circular, spiral, or other complex patterns, no linear hyperplane can separate the classes. Nonlinear decision boundaries are required.
The Kernel Trick: The Big Idea
Feature Mapping φ(x)
One solution: transform the data to a higher-dimensional space where it becomes linearly separable.
Example: For circular data in 2D, map $(x_1, x_2)$ to 3D:
$$\phi(x) = (x_1^2, \sqrt{2}x_1 x_2, x_2^2)$$
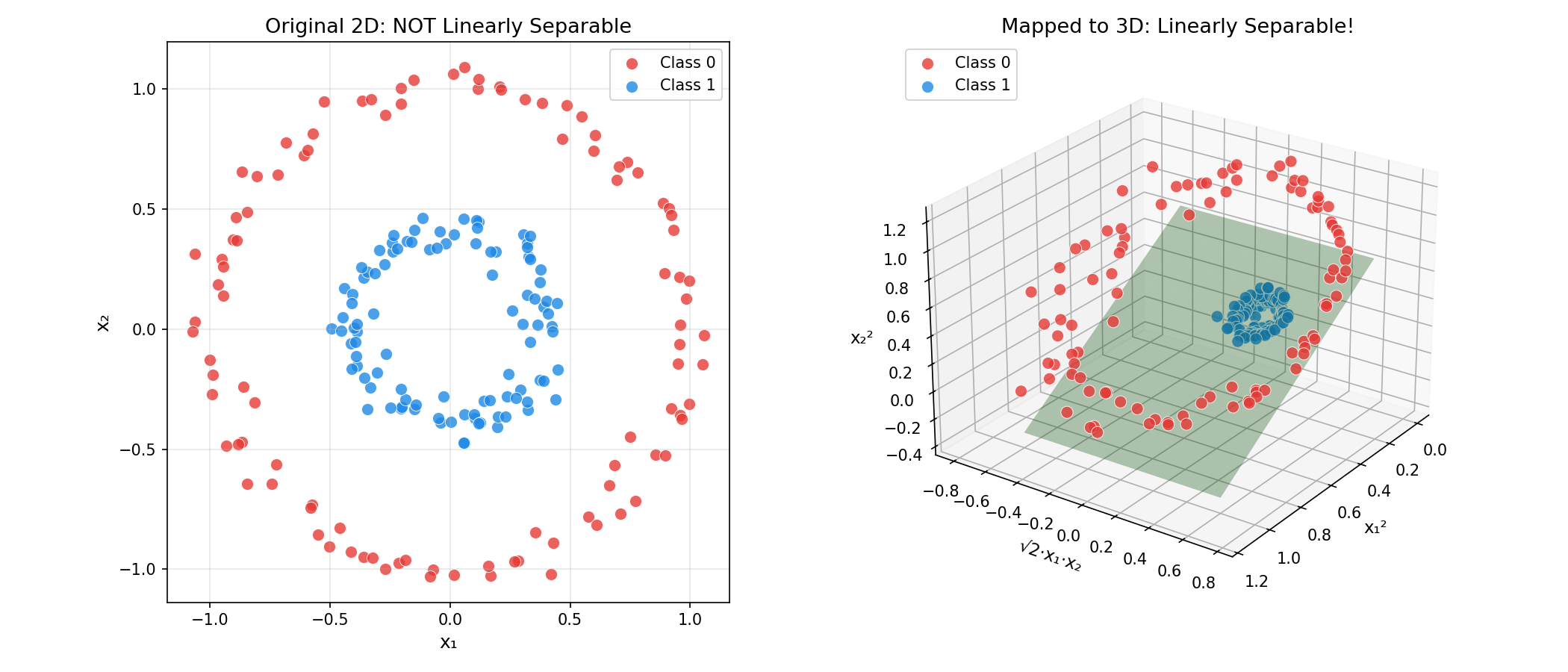
In this new space, the circular boundary becomes a flat plane!
The Computational Problem
But there’s a catch: if we map to a very high-dimensional space (or even infinite dimensions), computing $\phi(x)$ explicitly is expensive or impossible.
The Kernel Trick Solution
Recall from the dual formulation, the optimization only involves dot products $x_i^T x_j$.
Key insight: We can compute the dot product in the transformed space without explicitly computing $\phi(x)$:
$$K(x_i, x_j) = \phi(x_i)^T \phi(x_j)$$
This is the kernel function — it computes the inner product in high-dimensional space using only the original features!
Why this works: The kernel function is a “shortcut” that gives the same result as:
- Transform $x_i$ and $x_j$ to high dimensions
- Compute their dot product
But step 1 is skipped entirely!
Common Kernels
Linear Kernel
$$K(x_i, x_j) = x_i^T x_j$$
- What it does: No transformation, equivalent to standard SVM
- When to use: Data is already linearly separable
- Advantage: Fastest, least prone to overfitting
Polynomial Kernel
$$K(x_i, x_j) = (\gamma \cdot x_i^T x_j + r)^d$$
where $d$ is the degree, $r$ is the coefficient (coef0), and $\gamma$ scales the dot product.
Geometric intuition:
- $d = 2$: Can learn elliptical and parabolic boundaries
- $d = 3$: Can learn cubic curves
- Higher $d$: More complex shapes (but risk overfitting)
| Parameter | Effect |
|---|---|
degree | Polynomial degree — higher = more flexible |
coef0 | Shifts the polynomial (affects shape) |
gamma | Scales input features |
RBF (Radial Basis Function) Kernel
$$K(x_i, x_j) = \exp\left(-\gamma |x_i - x_j|^2\right)$$
Also called the Gaussian kernel. This is the most popular kernel!
Geometric intuition:
- Measures similarity between points
- Points close together → $K \approx 1$
- Points far apart → $K \approx 0$
The magic: RBF implicitly maps to an infinite-dimensional space! Yet we compute it with a simple formula.
The γ (Gamma) Parameter
| γ Value | Effect | Risk |
|---|---|---|
| High γ | Each point has very local influence | Overfitting (complex, wiggly boundary) |
| Low γ | Points influence far away | Underfitting (too smooth boundary) |
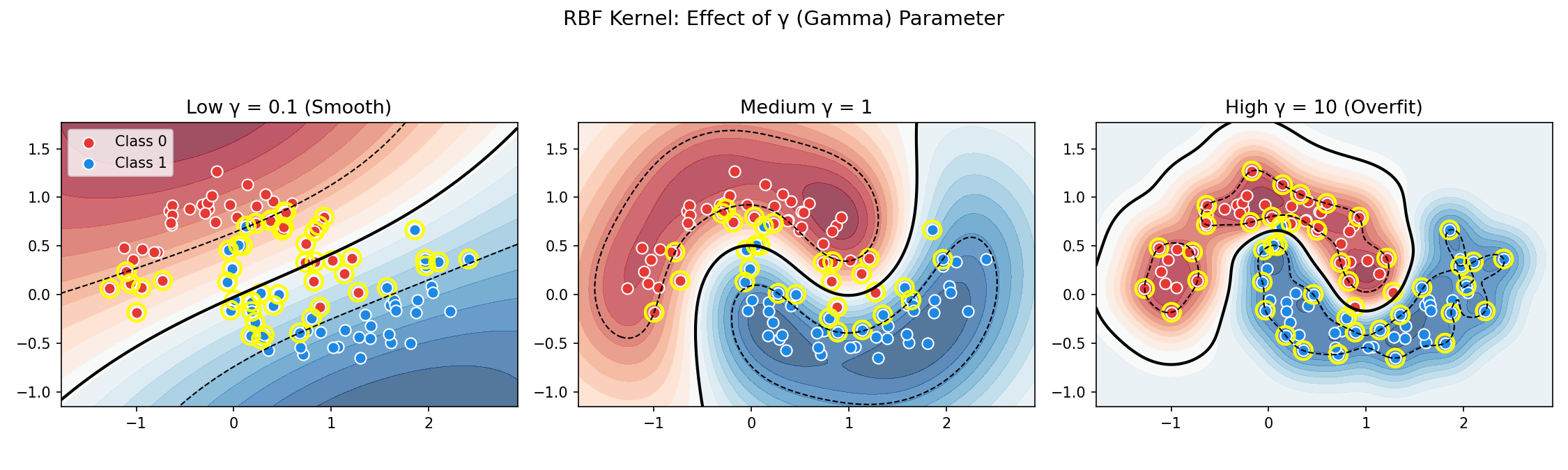
gamma='scale' without checking. Always visualize or cross-validate!Soft Margin SVM: Handling Noise
Why Hard Margin Fails
Real-world data is messy:
- Outliers that cross the margin
- Overlapping classes with no clean separation
- Noise that makes perfect separation impossible
Hard-margin SVM will either:
- Fail completely (no solution)
- Find a terrible boundary trying to fit outliers
The Solution: Allow Some Mistakes
Introduce slack variables $\xi_i \geq 0$ that measure how much each point violates the margin:
$$y_i(w^T x_i + b) \geq 1 - \xi_i$$
| $\xi_i$ Value | Meaning |
|---|---|
| $\xi_i = 0$ | Point is on or outside the margin (correct) |
| $0 < \xi_i < 1$ | Point is inside the margin but correct side |
| $\xi_i \geq 1$ | Point is misclassified |
The Soft-Margin Optimization Problem
Minimize:
$$\boxed{\frac{1}{2}|w|^2 + C \sum_{i=1}^{N} \xi_i}$$
Subject to:
- $y_i(w^T x_i + b) \geq 1 - \xi_i, \quad \forall i$
- $\xi_i \geq 0, \quad \forall i$
The objective balances:
- $\dfrac{1}{2}|w|^2$ — maximize margin (as before)
- $C \sum \xi_i$ — penalize violations
The C Parameter: Bias-Variance Tradeoff
$C$ controls how much we penalize margin violations:
| C Value | Margin | Errors | Behavior |
|---|---|---|---|
| Large C | Narrow | Few allowed | Low bias, high variance — fits training data tightly |
| Small C | Wide | More allowed | High bias, low variance — smoother, more generalizable |
Rule of thumb:
- Start with
C=1and adjust based on cross-validation - If overfitting → decrease C
- If underfitting → increase C
Soft-Margin Dual Problem
The dual becomes:
$$\max_\alpha \sum_{i=1}^{N} \alpha_i - \frac{1}{2} \sum_{i,j} \alpha_i \alpha_j y_i y_j K(x_i, x_j)$$
Subject to:
- $0 \leq \alpha_i \leq C, \quad \forall i$ (the box constraint!)
- $\sum_{i=1}^{N} \alpha_i y_i = 0$
Three Types of Points
After training, each point falls into one category:
| Condition | Type | Role |
|---|---|---|
| $\alpha_i = 0$ | Non-support vector | Far from boundary, doesn’t affect model |
| $0 < \alpha_i < C$ | Free support vector | Exactly on the margin |
| $\alpha_i = C$ | Bounded support vector | Inside margin or misclassified |
Code Practice
The following examples demonstrate kernels and soft margins in action.
Comparing Kernels on Circular Data
This example shows why kernel choice matters. The make_circles data creates concentric rings — impossible to separate linearly.
🐍 Python
| |
Output:
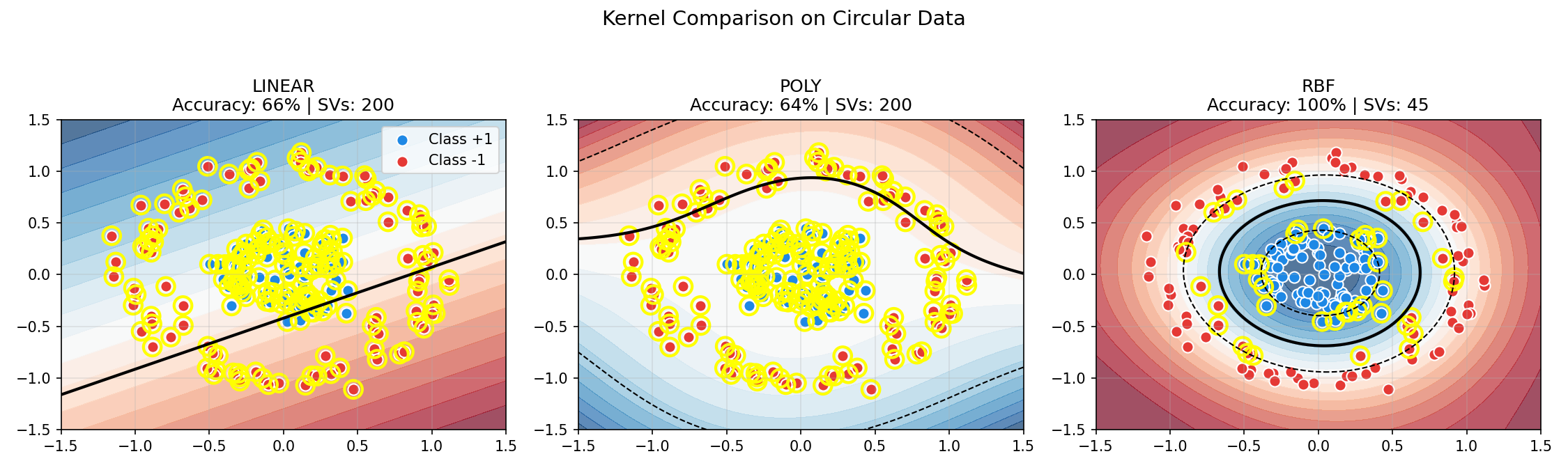
Key observations:
- Linear and Polynomial kernels fail on circular data (~65%)
- RBF achieves perfect separation (100%) by mapping to infinite dimensions
- More support vectors indicate a more complex decision boundary
Effect of C Parameter (Soft Margin)
The following visualization shows how C affects the decision boundary on noisy data:
🐍 Python
| |
Output:
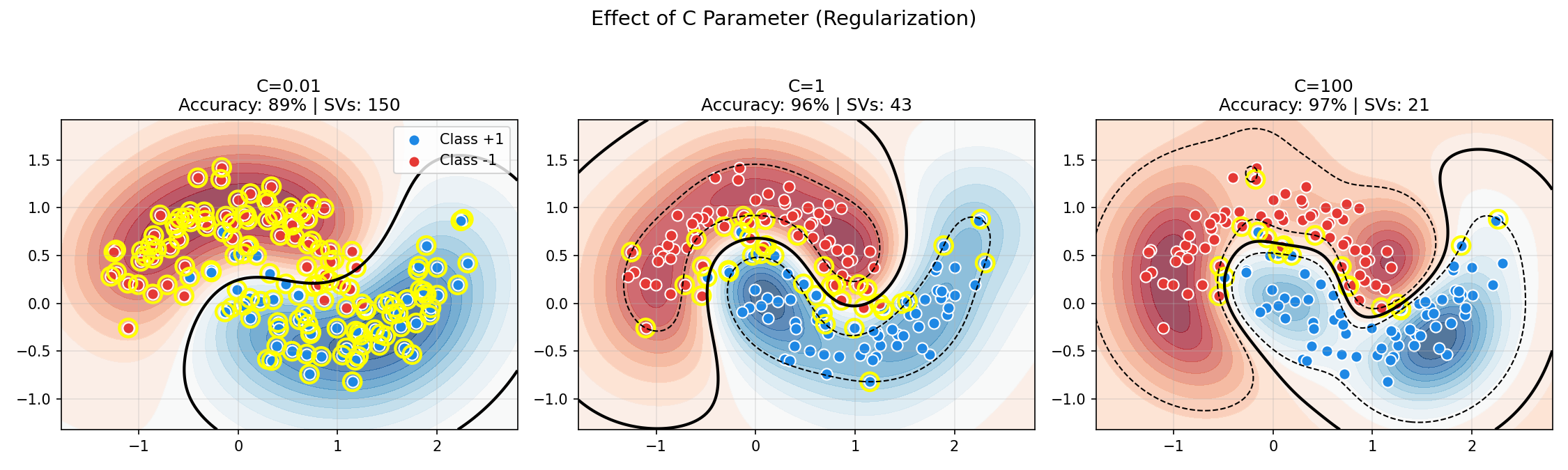
Observe the tradeoff:
- Low C (soft margin): Smooth boundary, many support vectors, tolerates misclassification
- High C (hard margin): Complex boundary, fewer support vectors, fits noise
- Balanced C: Best generalization on noisy data
Grid Search for Hyperparameters
Always use cross-validation to find optimal C and γ:
🐍 Python
| |
Output:
| |
RandomizedSearchCV.Deep Dive
Frequently Asked Questions
Q1: How do I choose between kernels?
Start with this decision flowchart:
| Data Characteristics | Recommended Kernel | Why |
|---|---|---|
| High-dim text/documents | Linear | Features are already expressive; linear is fast |
| Low-dim with clear curves | Polynomial (d=2-3) | Can fit ellipses, parabolas |
| Complex, unknown structure | RBF | Universal approximator |
| Very large dataset | Linear or Approx. RBF | RBF is $O(n^2)$ |
Q2: What does γ (gamma) really do?
γ controls the “reach” of each training point’s influence:
$$K(x_i, x_j) = \exp(-\gamma |x_i - x_j|^2)$$
| γ Value | Influence Radius | Decision Boundary | Risk |
|---|---|---|---|
| Very high (e.g., 100) | Tiny | Wiggly, fits every point | Severe overfitting |
| High (e.g., 10) | Small | Complex, detailed | Overfitting |
| Medium (e.g., 1) | Moderate | Balanced | - |
| Low (e.g., 0.1) | Large | Smooth | Underfitting |
| Very low (e.g., 0.01) | Huge | Nearly linear | Severe underfitting |
Q3: How do C and γ interact?
Both affect model complexity, but in different ways:
| Parameter | Controls | Too Low | Too High |
|---|---|---|---|
| C | Penalty for misclassification | Underfitting (too smooth) | Overfitting (fits outliers) |
| γ | Locality of influence | Underfitting (too smooth) | Overfitting (too complex) |
Practical tuning strategy:
- Fix
γ='scale'(sklearn default), tune C first - Then tune γ with best C
- Or use 2D grid search from the start:
- C: [0.1, 1, 10, 100, 1000]
- γ: [0.001, 0.01, 0.1, 1, 10]
Q4: Why is my SVM slow?
| Symptom | Cause | Solution |
|---|---|---|
| Training takes forever | Large N with RBF kernel | Use LinearSVC or reduce data |
| Many support vectors | C too small or data overlaps | Increase C or use linear kernel |
| Prediction is slow | Too many SVs | Reduce SVs via regularization |
RBF scaling: Training time is approximately $O(n^2)$ to $O(n^3)$. For >10,000 samples, consider:
LinearSVC(much faster)SGDClassifierwith kernel approximation- Subsampling the training data
Kernel Selection Guide
| Domain | Typical Kernel | Reason |
|---|---|---|
| Text Classification | Linear | TF-IDF features are high-dimensional and often linearly separable |
| Image Classification | RBF or CNN | Pixel relationships are complex |
| Genomics | Linear or String kernels | High-dimensional sparse features |
| Time Series | RBF or custom | Depends on the distance metric |
| Financial Data | RBF | Often has complex nonlinear patterns |
Hyperparameter Tuning Strategies
Coarse-to-Fine Search
Common Pitfalls
| Pitfall | Symptom | Solution |
|---|---|---|
| Not scaling features | Poor performance, sensitive to feature magnitude | Always use StandardScaler |
| Using RBF on linearly separable data | Slow training, similar accuracy to linear | Try linear kernel first |
| Overfitting with high C and γ | Great train accuracy, poor test accuracy | Use cross-validation, reduce C/γ |
| Default γ=‘scale’ on unnormalized data | Unpredictable results | Scale data first, then use defaults |
| Grid search on entire dataset | Optimistic bias | Use nested cross-validation |
When NOT to Use SVM
| Scenario | Better Alternative |
|---|---|
| Very large datasets (N > 100k) | Gradient boosting, neural networks |
| Need probability outputs | Logistic regression, calibrated SVM |
| Many noisy features | Random forest (automatic feature selection) |
| Online learning required | SGDClassifier |
| Interpretability critical | Decision trees, logistic regression |
Summary
| Concept | Key Points |
|---|---|
| Kernel Trick | Implicit high-dim mapping via $K(x_i, x_j)$ |
| RBF Kernel | $\exp(-\gamma|x_i - x_j|^2)$ — most flexible |
| Soft Margin | Allows $\xi_i$ slack for misclassifications |
| C Parameter | Tradeoff margin width vs errors |
References
- Boser, B. et al. (1992). “A Training Algorithm for Optimal Margin Classifiers”
- sklearn Kernel SVM Tutorial
- Schölkopf, B. & Smola, A. “Learning with Kernels”